|
|
Clonal Population Structure 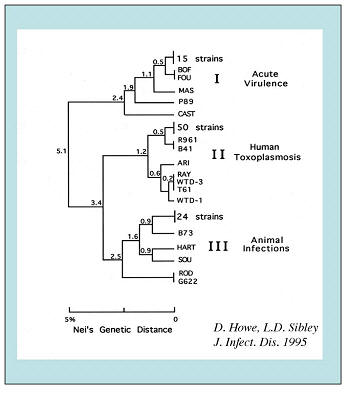 T.
gondii
has a highly unusual population genetic pattern consisting of three
widespread, clonal lineages known as types I, II and III
(Howe
and Sibley 1995).
Despite the presence of a meiotic stage in the
life
cycle
this clonal pattern persists in Nature, suggesting that much of
transmission is based on mitotic replication. The three lineages are
highly related, differing by only ~2% at the
nucleotide level. Studies of
genetic polymorphism reveal that at each locus there exist only two alleles,
indicating these three lineages arose from a single genetic recombination in the
environment
(Grigg et al.
2001; Su et al. 2003). T.
gondii
has a highly unusual population genetic pattern consisting of three
widespread, clonal lineages known as types I, II and III
(Howe
and Sibley 1995).
Despite the presence of a meiotic stage in the
life
cycle
this clonal pattern persists in Nature, suggesting that much of
transmission is based on mitotic replication. The three lineages are
highly related, differing by only ~2% at the
nucleotide level. Studies of
genetic polymorphism reveal that at each locus there exist only two alleles,
indicating these three lineages arose from a single genetic recombination in the
environment
(Grigg et al.
2001; Su et al. 2003).
|
|
|
The small genetic differences between strains likely underlie important biological differences. For example, type I strains are acutely lethal in mice (LD100 = 1 viable organism), and genetic linkage analysis has been used to analyze genes controlling this trait (Su et al. 2002). It remains uncertain to what extent the genotype of the parasite contributes to clinical severity in human toxoplasmosis (Boothroyd and Grigg 2002). Some studies indicate an increased prevalence of type I strains in human toxoplasmosis, although samples sizes are relatively small, geographic coverage is not uniform, and referral bias remains a problem. In most locations, type II strains are the most prevalent in human toxoplasmosis, while type III strains are largely found in animals. Further studies on the genotype of T. gondii strains from human infections, as well as animals, are needed from a wider geographic range. DNA polymorphisms can easily be used to genotype new isolates. |
Department of Molecular Microbiology
Washington University School of Medicine
St. Louis, MO USA