|
 |
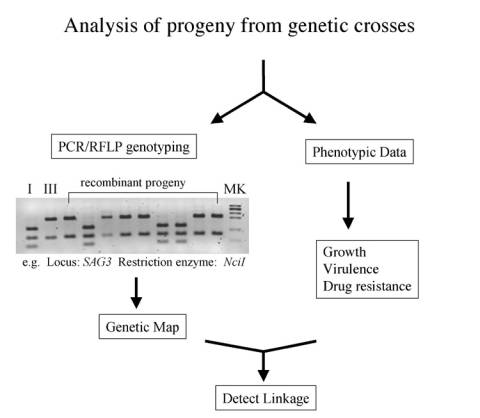
One of the major phenotypic differences between T. gondii strains is their acute virulence in the mouse. To analyze the genetic basis of this trait, a genetic cross was performed between the virulent type I strain GT-1 (Dubey 1980) and nonvirulent the type III strain CEP. This project was conducted jointly between the laboratories of Dr. J.P. Dubey at the USDA, Dr. Jim Ajioka at Cambridge University and Dr. David Sibley at Washington University. |
|||||||||||||||
|
Two parallel crosses were performed as shown above. Progeny were selected by segregation between the drug resistance markers AraA, SNF, and FUDR. The number of clones analyzed from each cross are summarized in Table 1. Remarkably, all randomly selected progeny were recombinants, despite previous estimates that self-mating and out-crossing should be equally probable. Table 1 Results of IxIII crosses
A total of 26 progeny were analyzed using 53 PCR-RFLP markers. When combined with the analysis of a IIxIII genetic cross, these data were used to generate improved linkage maps using MapMaker EXP with a minimum LOD score of 3.0 (Su et al. 2002). Quantitative trait locus (QTL) mapping was used to map loci responsible for virulence and this led to the identification of a primary locus controlling virulence on chromosome VII (Su et al. 2002). The data from analysis of these progeny has been incorporated into the Toxoplasma Genome Mapping Database. |
||||||||||||||||
Department of Molecular Microbiology
Washington University School of Medicine
St. Louis, MO USA